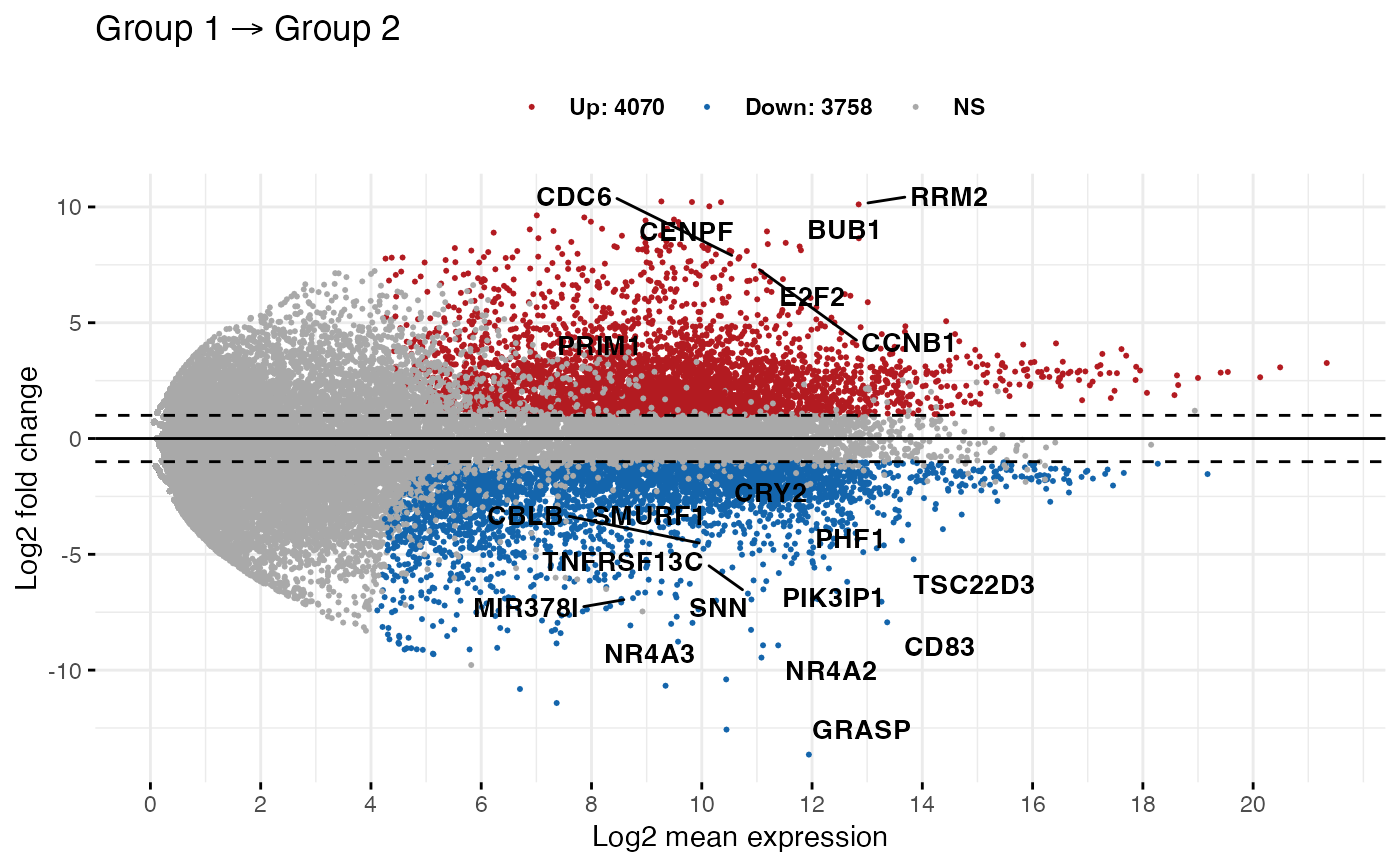
MD plot showing the log-fold change and average abundance of each gene.... | Download Scientific Diagram
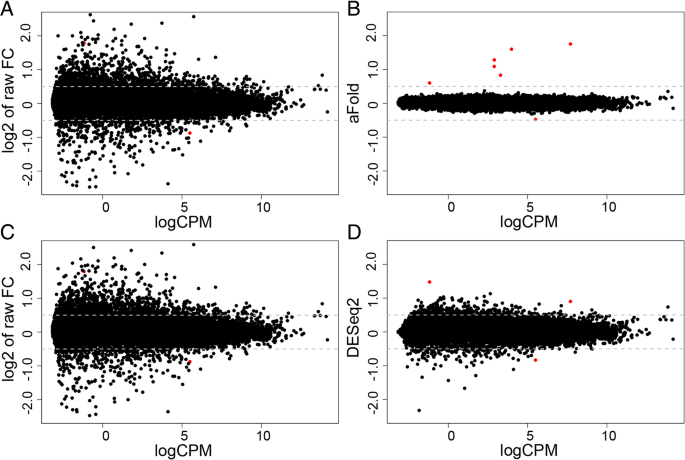
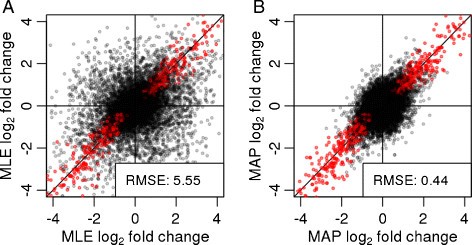
aFold – using polynomial uncertainty modelling for differential gene expression estimation from RNA sequencing data | BMC Genomics | Full Text

Tuning Transcriptional Regulation through Signaling: A Predictive Theory of Allosteric Induction - ScienceDirect

Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram

Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram
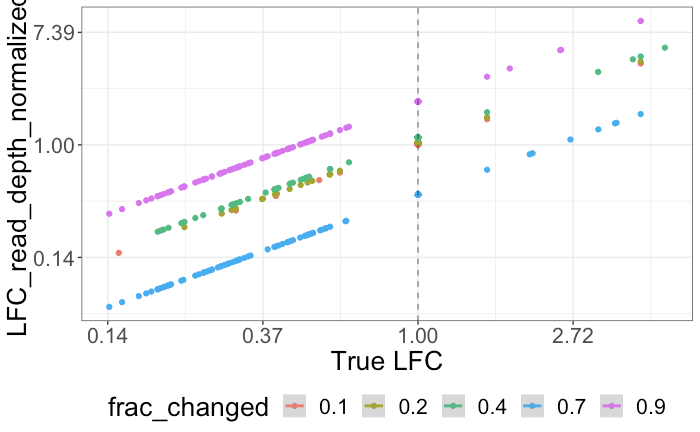
A comparison of per sample global scaling and per gene normalization methods for differential expression analysis of RNA-seq data

Transite: A Computational Motif-Based Analysis Platform That Identifies RNA-Binding Proteins Modulating Changes in Gene Expression - ScienceDirect
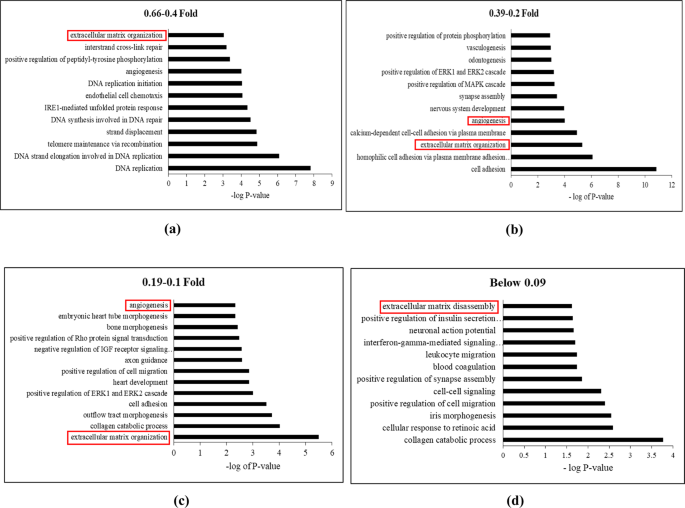
Transcriptomic Profiling of Adipose Derived Stem Cells Undergoing Osteogenesis by RNA-Seq | Scientific Reports